| Navigation: | Site Map | Copyright | Contact us | Acknowledgements | | |
|
Glu/Leu/Phe/Val dehydrogenases family |
|
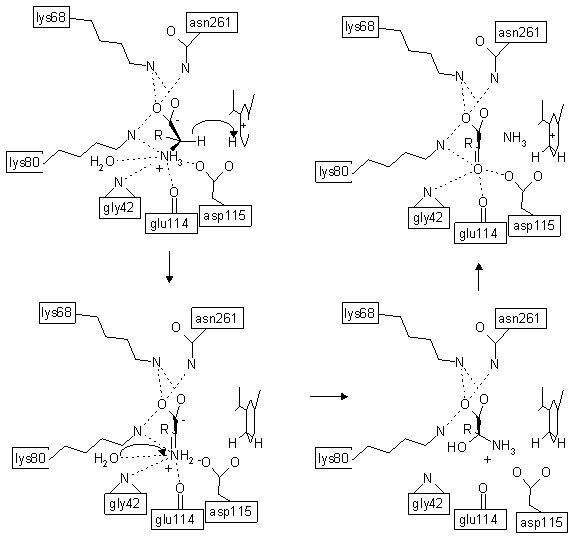
About Glu/Leu/Phe/Val dehydrogenases familyThe Glu/Leu/Phe/Val dehydrogenase family catalyses the oxidative deamination of amino acid to the appropriate ketone with the concomitant reduction of NAD+ to NADH. For example leucine is converted to 2-ketoisocaproate, glutamate is converted to 2-oxoglutarate etc. The Glu/Leu/Phe/Val dehydrogenase family is belongs to the amino acid dehydrogenase family which belongs to the Oxidoreductases Acting on CH-NH2 Group, according to the MESH classification.The sequence similarity across all members of the Glu/Leu/Phe/Val dehydrogenase family is 18-20%. Many enzymes from this family have considerable potential for commercial use [Bommarius, A.S., Schwarm, M. and Drauz, K. (1998) Biocatalysis to amino acid-based chiral pharmaceuticals - examples and perspectives. J. of Molecular Catalysis B: Enzymatic. 5, 1-11.]. For example, LeuDH has been used to production of -hydroxyvaline, an intermediate in the synthesis of the antibiotic Tigemonam [Hanson, R.L., Singh, J., Kissick, T.P., Patel, R.N., Szarka, L.J. and Mueller, R.H. (1990). Synthesis of L- -hydrohyvaline from -keto- -hydroxyisovalerate using LeuDH from Bacillus species. Bioorg. Chem. 18, 116-130.], or for production of L-tert-leucine, and as building blocks for pharmaceutically active compounds of drugs against different tumors, rheumatic arthritis and AIDS. The quaternary structure of Glu/Leu/Phe/Val dehydrogenases is always multimeric. The most common ones are dimeric structures for valine dehydrogenases, hexameric structures for glutamate dehydrogenases and octameric structures for leucine and phenylalanine dehydrogenases. Monomers from the Glu/Leu/Phe/Val dehydrogenase family consist from two domains, connected by flexible hinge regions. These two domains are separated by a deep cleft. The first domain consists of about 130 N-terminal residues and about 30 C-terminal residues and is composed of a mixed parallel/antiparallel β-sheet, flanked from both sides by α-helices. The second domain is the classical dinucleotide-binding Rossmann domain. The hinge region can be considered as a third domain of about 10 amino acid residues which can be selected between C-terminal end of Rossmann domain and the second polypeptide chain from the first domain. Structural studies on enzymes from the Glu/Leu/Phe/Val dehydrogenase family have suggested that substrate binding and product release strictly require an open form of these enzymes in which the enzymes two domains are widely separated by a deep cleft. In contrast for hydride transfer in which the two domains undergo a major conformational rearrangement which results in closure of the cleft during catalysis. During this rearrangement two domain can rotate up to 30 degree. The catalytic mechanism of Glu/Leu/Phe/Val dehydrogenases familyThe catalytic mechanism for Glu/Leu/Phe/Val dehydrogenases is conserved within this family. This catalytic mechanism is well studied for glutamate dehydrogenase, leucine dehydrogenase and phenylalanine dehydrogenase.The substrate is bound by an open form of the enzyme, and then the protein globule of Glu/Leu/Phe/Val dehydrogenase is closed and traps the substrate completely between two monomers, so the substrate is completely buried from the solvent. Only one water molecule, which is involved in catalysis, is trapped near the substrate. The main feature of this catalytic act is the large conformational changes of the Glu/Leu/Phe/Val dehydrogenase molecule. The relative rotation of subunits is up to 30 degree. The role of such a large shift is unclear. From one hand it can be either an adjustment of the active site to the catalytic needs or the "forced" catalytic act. All catalytic process can be split up into tree principal steps. The three steps for the structure of the Leucine dehydrogenase are the following:
|
| dehydrogenases.com: Copyright 2007 by dehydrogenases.com |